Summary
Research interests

Pr Vladimir Torbeev
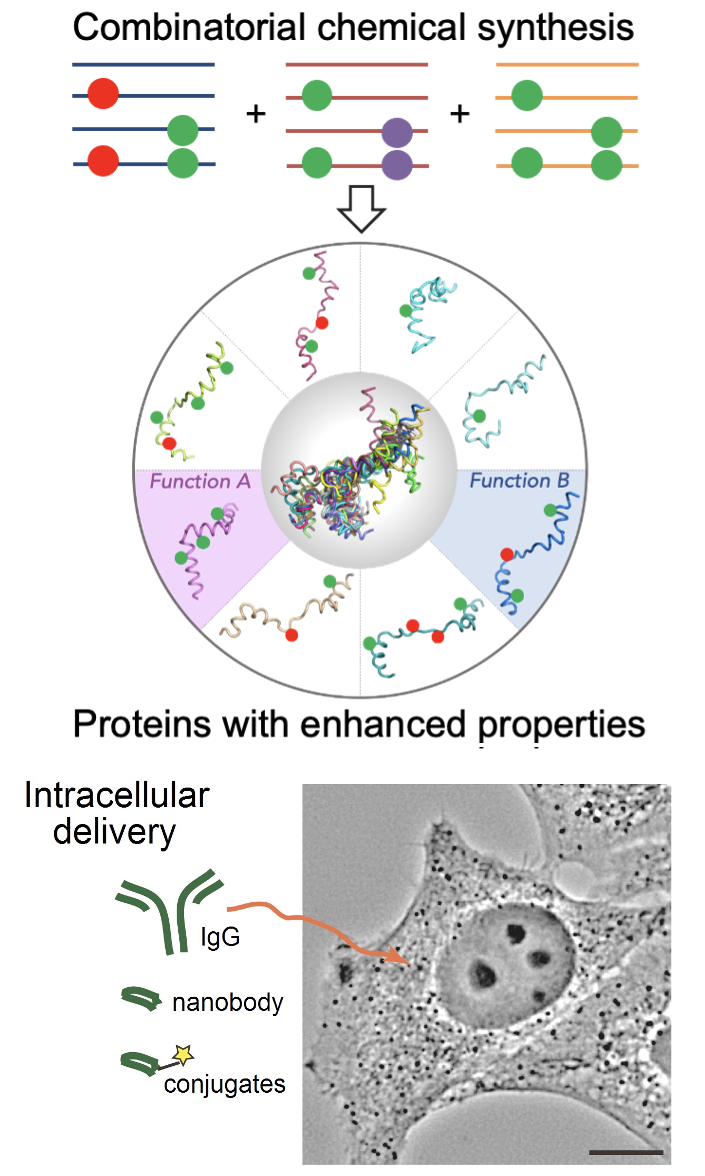
The research team “Biosystems Chemistry” led by Prof. Vladimir Torbeev specializes in the synthesis of proteins for biophysical and biological studies. By combination of solid-phase peptide synthesis, recombinant protein expression and chemoselective ligation chemistries (e.g. native chemical ligation), unique proteins can be produced possessing combinations of post-translational modifications, labels (isotopic, fluorescent and others) and various non-canonical amino acids. Precisely positioned modifications facilitate structure-function, biophysical and biological studies. In the past years, we synthesized and studied covalently tethered soluble amyloid oligomer, modified variants of activation domains of transcriptional coregulators p160 and CBP/p300 as well as transactivation domain of tumor suppressor p53. To characterize the function of proteins in cells, our laboratory is developing approaches for intracellular delivery of biomolecules (supervised by Dr. Guy Zuber). Functional studies and biological activity of synthetic proteins are supervised by Dr. Edwige Voisset.
Intrinsically disordered proteins. Nearly one third of eukaryotic proteome is composed of intrinsically disordered proteins or proteins that contain intrinsically disordered domains. Such proteins play highly important functional roles in cellular signaling, regulation and recognition. Furthermore, many disease-related proteins are intrinsically disordered. We would like to better understand the mechanisms of molecular recognition and complex formation that involve such proteins, their role in protein interaction networks and to elaborate approaches for modulating their functions inside cells.
Protein aggregation. Protein-misfolding diseases (Alzheimer’s, Parkinson’s diseases and others) affect millions of people worldwide, however, the current understanding of these disorders is incomplete to develop efficient treatments. In these diseases, folded or unstructured proteins undergo conformational isomerization (misfolding) and self-assembly into toxic oligomers and amyloid fibrils. We aim at dissecting the molecular details of such molecular interconversions and identifying molecular targets for designing new drugs and diagnostics.
Protein design. Chemical synthesis of proteins is highly useful for the design of proteins with unique structures and functions. Non-canonical building blocks (D-amino acids, fluorinated residues, thiotyrosine) enable to modulate protein conformation, structure or chemical reactivity. To enable a highly efficient synthesis of new proteins we are designing protein catalysts to accelerate the ligation of synthetic and recombinant protein segments. Such artificial ligases will be particularly useful for the synthesis of protein libraries.
Intracellular chemical biology probes. The intracellular delivery of nucleic acids or proteins capable to interfere with cellular signalling in a targeted way is a key for being able to modulate biological processes. Existing nucleic acid vectorization methods are proved sufficiently effective for vaccination or the treatment of certain rare diseases. Our aim is to contribute to developing new methods for transferring biomolecules inside cells. In particular, we are working on the direct transfer of proteins and chemical biology probes to track and localize target proteins in living cells in real time and at high resolution.
Contact
Pr Vladimir Torbeev
Funding

Team members
- Bureau : D321
- mohamed.banni[at]unistra.fr
- Bureau : D439
- chiper[at]unistra.fr
Hanss Victor
Jouin Alexis
Négroni Luc
- Bureau : D321
- lnegroni[at]unistra.fr
Ofner Shalene
Torbeev Vladimir
- Bureau : D322-3
- torbeev[at]unistra.fr
Trostanetskaia Anastasiia
Voisset Edwige
- Bureau : D322-1
- voisset[at]unistra.fr
Yousef Mo’ath
- Bureau : D321
- Bureau : D306
- zuber[at]unistra.fr
Publications
Iesu L, Sai M, Torbeev V, Kieffer B, Pelta J, Cressiot B (2025). Single-molecule nanopore sensing of proline cis/trans amide isomers. Chem Sci 16(22):9730-9738. doi: 10.1039/d5sc01156f.
Bachelart T, Kumar S, Jouin A, Yousef M, Kieffer B, Torbeev V (2024). Design, Synthesis and Catalytic Activity of Protein Containing Thiotyrosine as an Active Site Residue. Chembiochem 17:e202400148. doi: 10.1002/cbic.202400148.
Grelich-Mucha M, Bachelart T, Torbeev V, Ożga K, Berlicki Ł, Olesiak-Bańska J (2024). Amyloid engineering - how terminal capping modifies morphology and secondary structure of supramolecular peptide aggregates. Biomater Sci 12(6):1590-1602. doi: 10.1039/d3bm01641b.
Aucagne V, Burlina F, Melnyk O, Torbeev V (2021). Des protéines de synthèse taillées sur mesure pour investiguer le vivant. L’actualité chimique 468:26-28.
Sinnaeve D, Ben Bouzayene A, Ottoy E, Hofman GJ, Erdmann E, Linclau B, Kuprov I, Martins JC, Torbeev V, Kieffer B (2021). Fluorine NMR study of proline-rich sequences using fluoroprolines. Magn Reson (Gott) 2(2):795-813. doi: 10.5194/mr-2-795-2021.
Grelich-Mucha M, Garcia AM, Torbeev V, Ożga K, Berlicki Ł, Olesiak-Bańska J (2021). Autofluorescence of Amyloids Determined by Enantiomeric Composition of Peptides. J Phys Chem B 125(21):5502-5510. doi: 10.1021/acs.jpcb.1c00808.
Naudin EA, McEwen AG, Tan SK, Poussin-Courmontagne P, Schmitt JL, Birck C, DeGrado WF, Torbeev V (2021). Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of α-Helix As Oxyanion-Binding Site. J Am Chem Soc 143(9):3330-3339. doi: 10.1021/jacs.0c10053.
Bauer V, Schmidtgall B, Gógl G, Dolenc J, Osz J, Nominé Y, Kostmann C, Cousido-Siah A, Mitschler A, Rochel N, Travé G, Kieffer B, Torbeev V (2020). Conformational editing of intrinsically disordered protein by α-methylation. Chem Sci 12(3):1080-1089. doi: 10.1039/d0sc04482b.
Garcia AM, Giorgiutti C, El Khoury Y, Bauer V, Spiegelhalter C, Leize-Wagner E, Hellwig P, Potier N, Torbeev V (2020). Aggregation and Amyloidogenicity of the Nuclear Coactivator Binding Domain of CREB-Binding Protein. Chemistry 26(44):9889-9899. doi: 10.1002/chem.202001847.
Torbeev V, Kent SBH (2020). Chemical synthesis of an enzyme containing an artificial catalytic apparatus. Australian J Chem 73:321-326
Torbeev V (2020). Illuminating Voltage Sensor Paddling in Different Membrane Milieu. Biophys J 118(4):781-782. doi: 10.1016/j.bpj.2019.09.032.
Vergauwe RMA, Thomas A, Nagarajan K, Shalabney A, George J, Chervy T, Seidel M, Devaux E, Torbeev V*, Ebbesen TW* (2019). Modification of Enzyme Activity by Vibrational Strong Coupling of Water. Angew Chem Int Ed Engl 58(43):15324-15328. doi: 10.1002/anie.201908876.
Baral A, Asokan A, Bauer V, Kieffer B, Torbeev V (2019). Chemical synthesis of transactivation domain (TAD) of tumor suppressor protein p53 by native chemical ligation of three peptide segments. Tetrahedron 75:703-708. doi: 10.1016/j.tet.2018.11.074.
Boehringer R, Kieffer B, Torbeev V (2018). Total chemical synthesis and biophysical properties of a designed soluble 24 kDa amyloid analogue. Chem Sci 9(25):5594-5599. doi: 10.1039/c8sc01790e.
Schmidtgall B, Chaloin O, Bauer V, Sumyk M, Birck C, Torbeev V (2017). Dissecting mechanism of coupled folding and binding of an intrinsically disordered protein by chemical synthesis of conformationally constrained analogues. Chem Commun (Camb) 53(53):7369-7372. doi: 10.1039/c7cc02276j.
Ruiz J, Boehringer R, Grogg M, Raya J, Schirer A, Crucifix C, Hellwig P, Schultz P, Torbeev V (2016). Covalent Tethering and Residues with Bulky Hydrophobic Side Chains Enable Self-Assembly of Distinct Amyloid Structures. Chembiochem 17(23):2274-2285. doi: 10.1002/cbic.201600440.
Vergauwe RMA, George J, Chervy T, Hutchison JA, Shalabney A, Torbeev VY, Ebbesen TW (2016). Quantum Strong Coupling with Protein Vibrational Modes. J Phys Chem Lett 7(20):4159-4164. doi: 10.1021/acs.jpclett.6b01869.
Torbeev V, Grogg M, Ruiz J, Boehringer R, Schirer A, Hellwig P, Jeschke G, Hilvert D (2016). Chiral recognition in amyloid fiber growth. J Pept Sci 22(5):290-304. doi: 10.1002/psc.2861.

